The advent of next-generation DNA sequencing is accelerating our discovery of rare gene variants that may represent causative mutations in speech and language disorders. Computer-based prediction methods can provide some insight into the potential consequences of these variants, but experimental techniques in cellular systems are crucial for confirming whether they are truly causal and for enabling a deeper understanding of their biological significance.
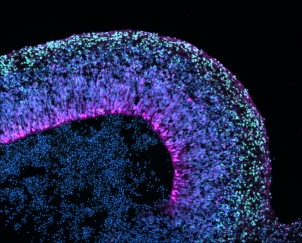
Our methods test the impact of gene variants on the properties and functions of encoded proteins through the genetic manipulation of human cells that we can grow in the laboratory. We assess the effects on expression level, protein stability, intracellular localisation, interaction of the protein with other molecules, as well as using specific assays of protein function. Over the past few years, we have used these techniques to demonstrate the aetiological impact of mutations in key regulatory genes, including FOXP2, FOXP1, TBR1, BCL11A, and CHD3. In our latest studies (not yet published), we are working with gene-editing (CRISPR/Cas9) and human stem cells, differentiating them into distinct types of human neurons and/or brain organoids. These newer cell-based models hold great promise for deciphering how mutations in FOXP2, and other language-related genes, affect aspects of neuronal development such as proliferation, migration, neurite outgrowth, and plasticity.
Example publications:
Snijders Blok, L. et al. (2021). Heterozygous variants that disturb the transcriptional repressor activity of FOXP4 cause a developmental disorder with speech/language delays and multiple congenital abnormalities. Genetics in Medicine, 23.3: 534-542. doi:10.1038/s41436-020-01016-6. [pdf]
Den Hoed, Joery, et al. (2021). Mutation-specific pathophysiological mechanisms define different neurodevelopmental disorders associated with SATB1 dysfunction. The American Journal of Human Genetics 108.2: 346-356. doi:10.1016/j.ajhg.2021.01.007. [pdf]
Den Hoed, J., et al. (2018). Functional characterization of TBR1 variants in neurodevelopmental disorder. Scientific Reports, 8: 14279. doi:10.1038/s41598-018-32053-6. [pdf]
Sollis, E., et al. (2017). Equivalent missense variant in the FOXP2 and FOXP1 transcription factors causes distinct neurodevelopmental disorders. Human Mutation, 38(11), 1542-1554. doi:10.1002/humu.23303. [pdf]
Dias, C., Estruch, S.B., et al. (2016). BCL11A haploinsufficiency causes an intellectual disability syndrome and dysregulates transcription. The American Journal of Human Genetics, 99(2), 253-274. doi:10.1016/j.ajhg.2016.05.030. [pdf]
Share this page